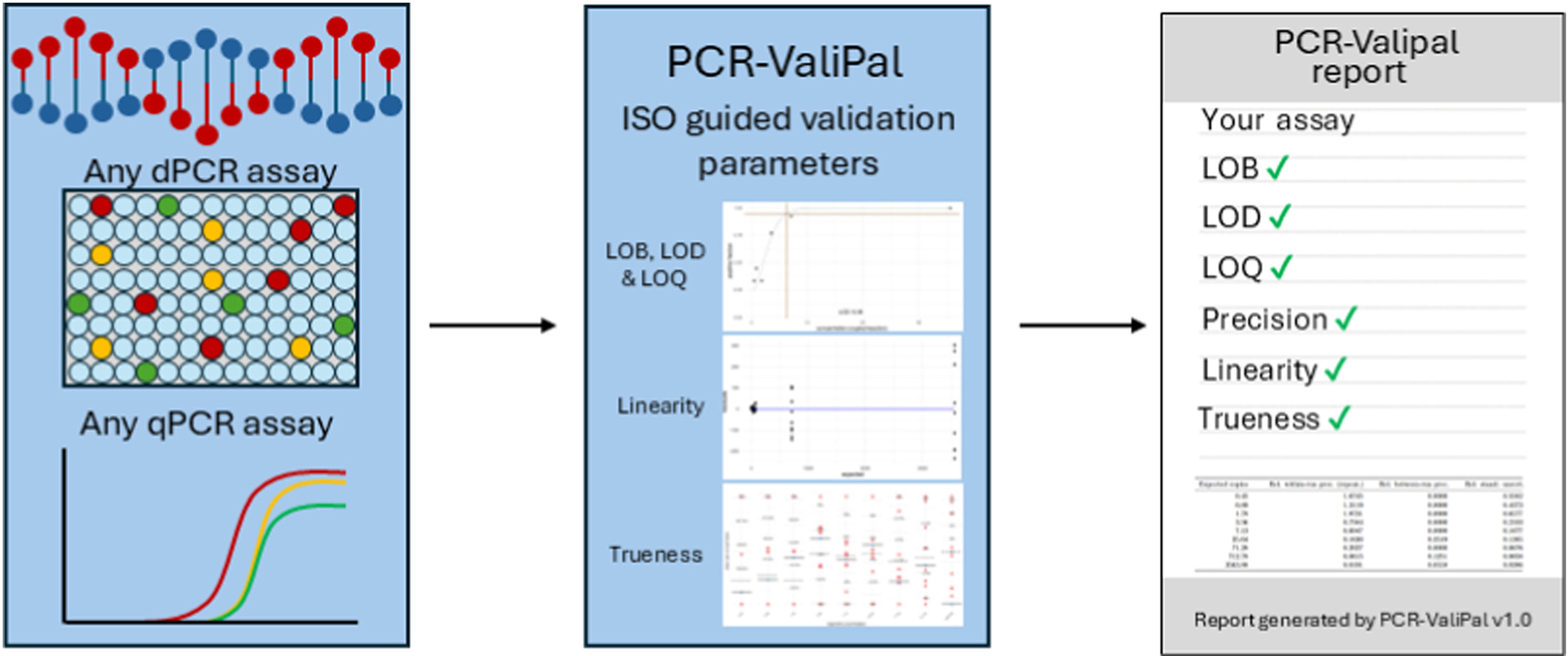
PCR ValiPal is a web-based application designed to standardize and automate the validation of PCR assays (qPCR and digital PCR). PCR ValiPal evaluates all key parameters required for PCR validation,
including: Limit of blank (LOB); Limit of detection (LOD); Limit of quantification (LOQ); Precision (repeatability & intermediate precision); Trueness/bias; Linearity
Featured Publications
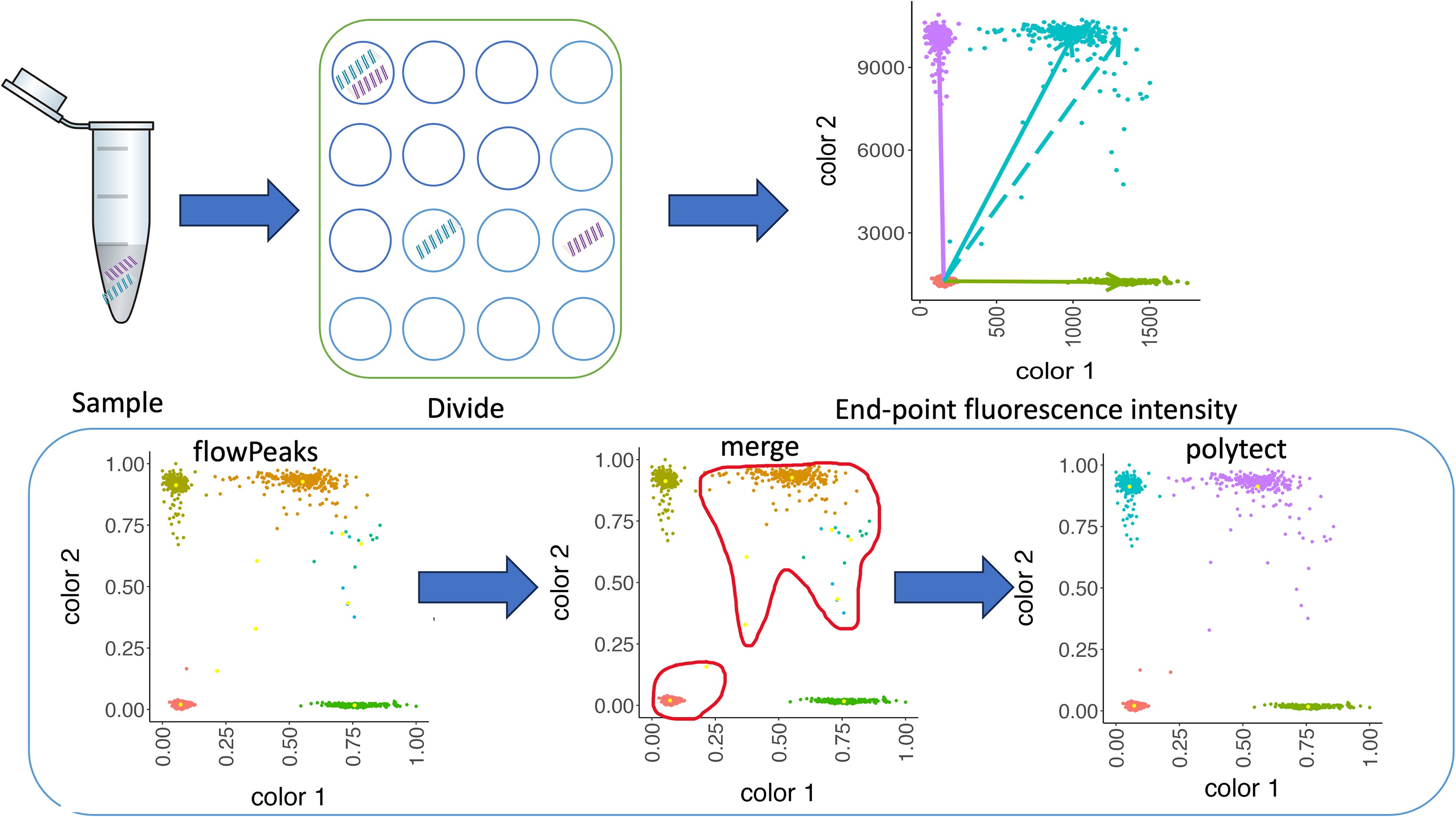
Polytect Builds on existing clustering methods, refining results by merging and labeling clusters based on prior knowledge of cluster center locations. Polytect outperforms other methods in both empirical and simulated data, effectively merging excess clusters and identifying empty clusters. The method is available as an R package and a Shiny app for easy use.
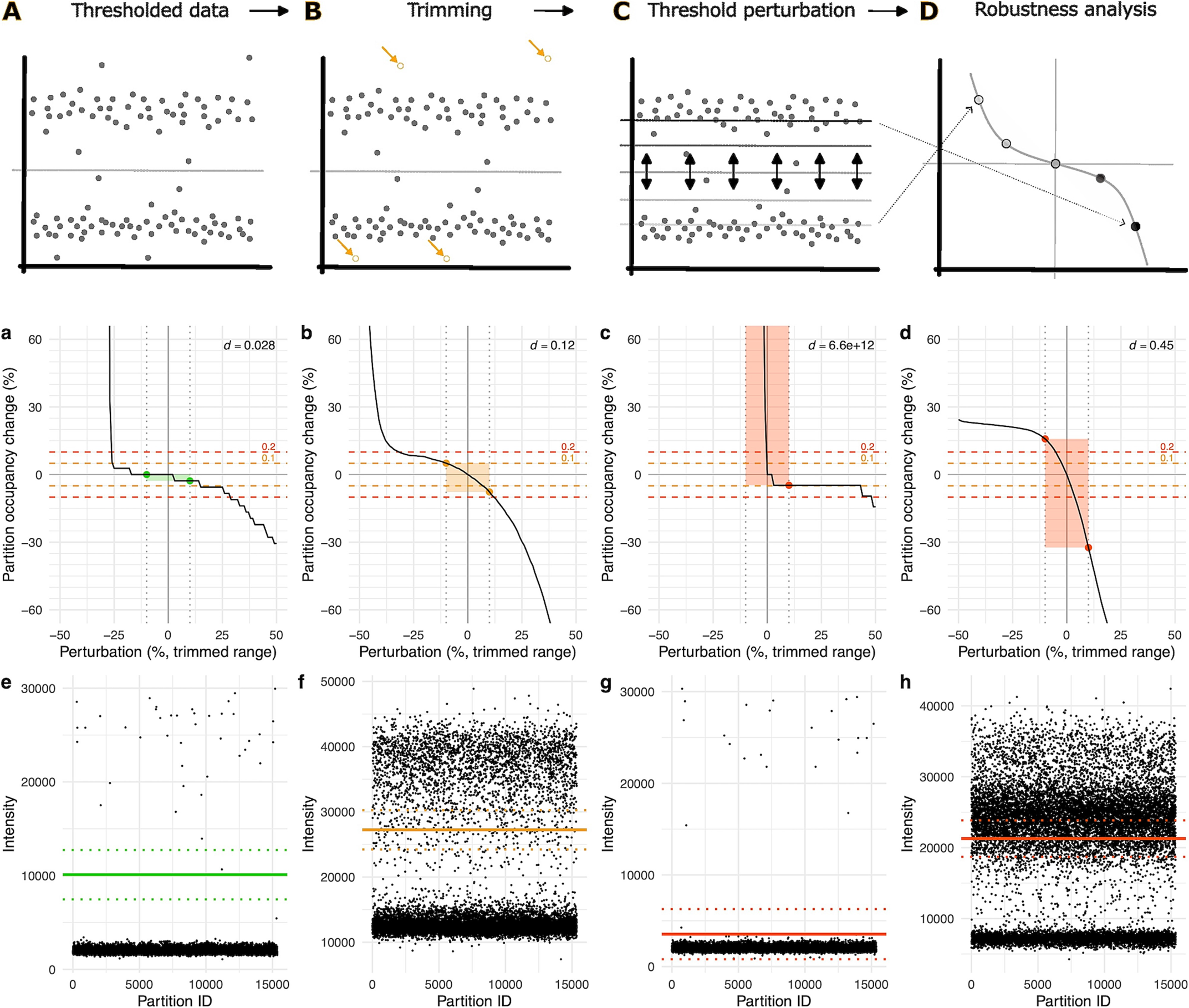
DipcensR provides automates the assessment of partition classification robustness, reducing the need for extensive manual review.
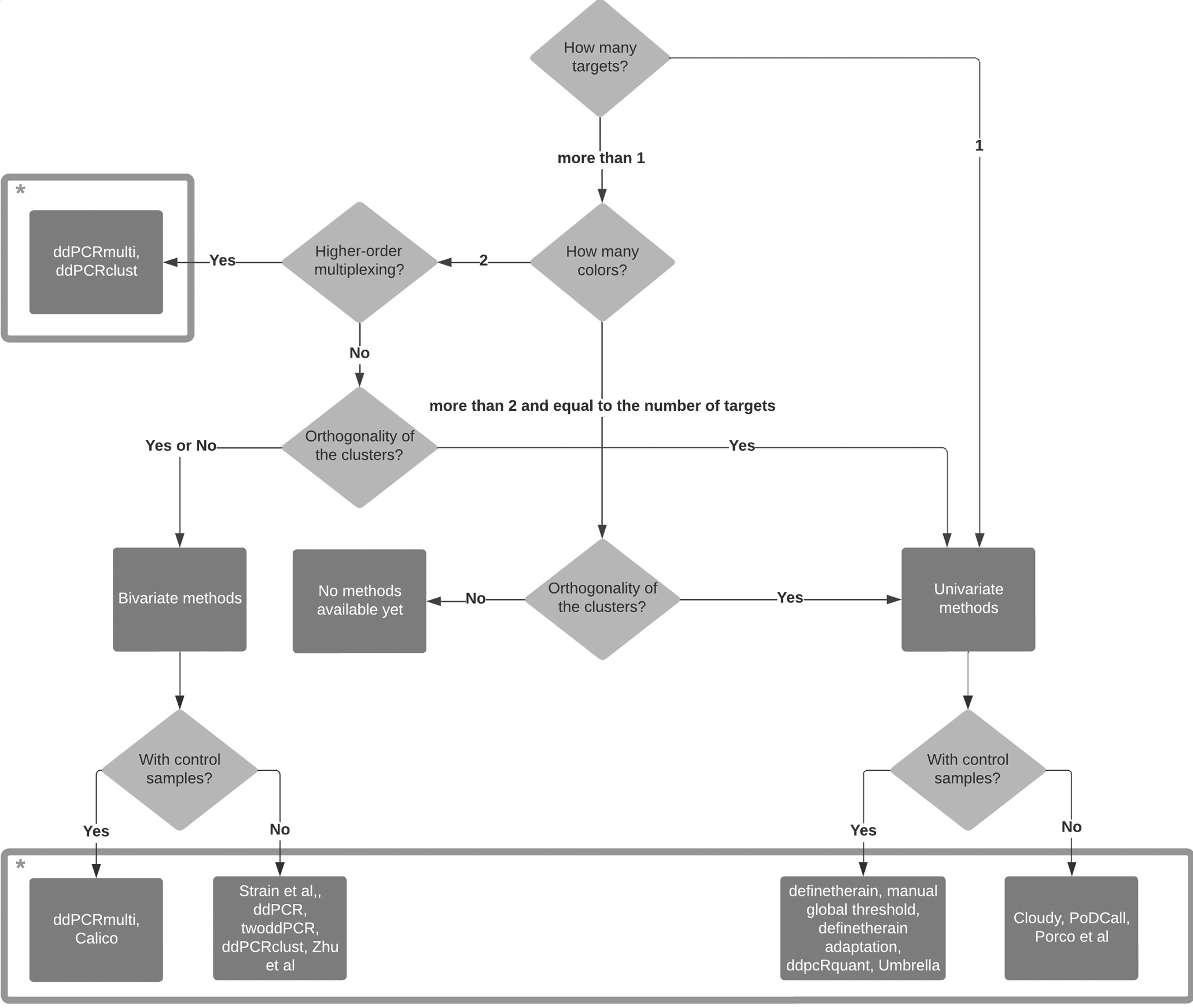
Summary of all available digital PCR partition classification approaches and the challenges
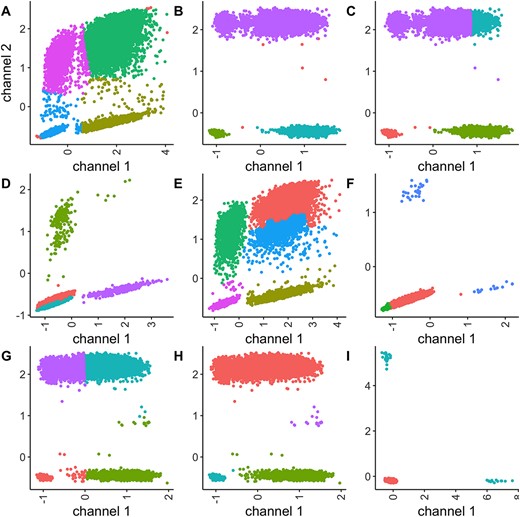
This paper evaluates how well different clustering algorithms classify partitions in duplex digital PCR experiments, using both real and simulated data.
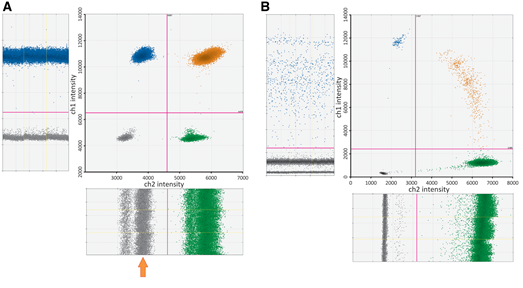
Minimum Information for Publication of Quantitative Digital PCR Experiments for 2020
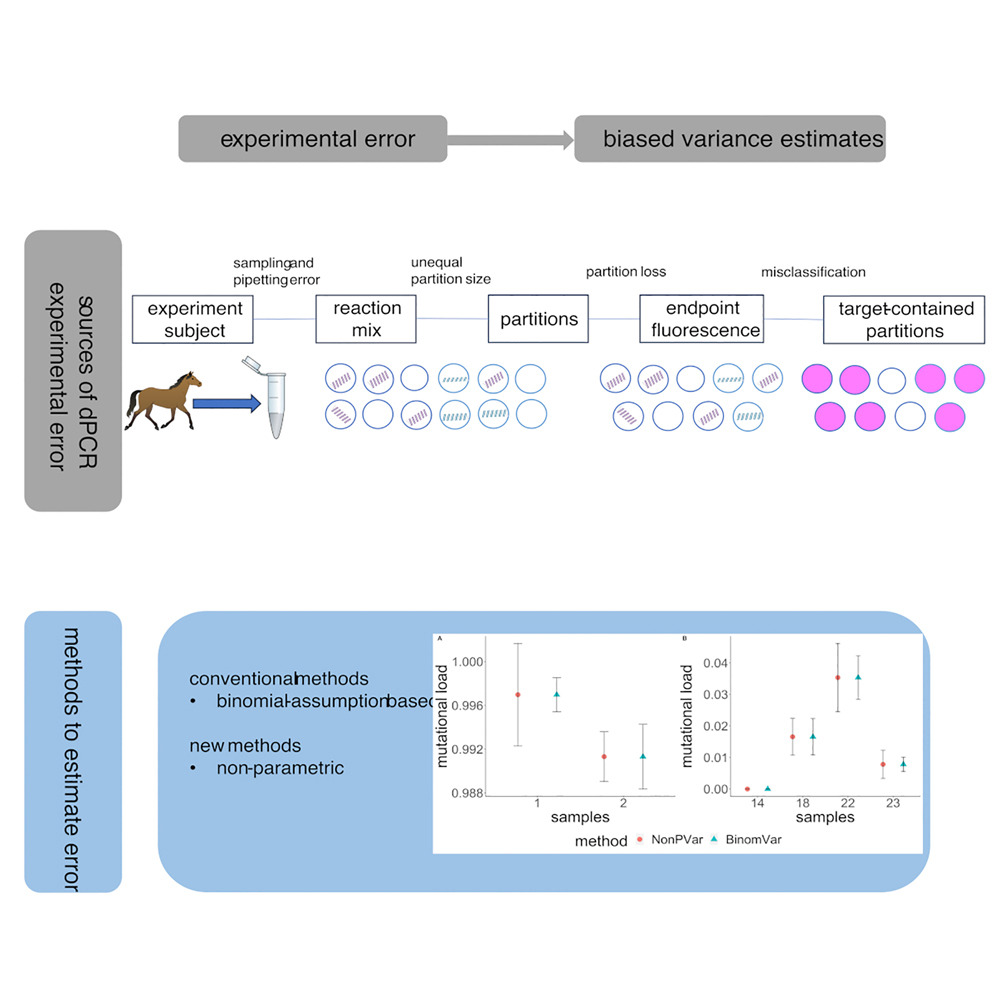
This paper introduces two new, flexible methods (NonPVar and BinomVar) to estimate variance and confidence intervals for digital PCR readouts, including complex quantities like CNV and fractional abundance. An R Shiny app is provided to make them easy to apply in practice.